Density-Based Spatial Clustering of Applications with Noise
K-Means is an efficient clustering algorithm that performs well when the number of clusters, \(K\), is appropriately specified. In contrast, Density-Based Spatial Clustering of Applications with Noise (DBSCAN) is a density-based algorithm that groups points according to their local density that enables the detection of clusters of arbitrary shapes and the identification of noise points.
To estimate density, DBSCAN relies on two key parameters: \(\varepsilon\) (or eps) and minPts. The former defines the maximum radius of a neighborhood around a point, while the latter specifies the minimum number of points required within this neighborhood for a point to be considered a core point. Based on these parameters, three types of points are defined:
-
Core Point: A point that has at least minPts neighbors (including itself) within its eps neighborhood. Core points represent dense regions that form the backbone of clusters.
-
Border Point: A point with fewer than minPts neighbors in its eps neighborhood, yet lies within the \textit{eps} neighborhood of a core point.
-
Noise Point: A point that is neither a core point nor a border point. Such points are considered outliers and do not belong to any cluster.
The algorithm begins with an unvisited data point, referred to as the point of interest. Then, It identifies all points within the eps distance of this point. If the number of neighboring points is less than minPts, the point is temporarily labeled as noise (it may later become a border point if it falls within the eps neighborhood of a core point). Otherwise, the point is identified as a c ore point, and a new cluster is initiated.
DBSCAN then recursively expands this cluster by adding all density-reachable points, i.e., all core points and their connected border points. This expansion continues by iterating through each newly added core point and including its neighbors if they also satisfy the core point condition or lie within the eps neighborhood of an existing core point. The process repeats until all points have been visited and assigned a label, either as part of a cluster or as noise.
Implementation
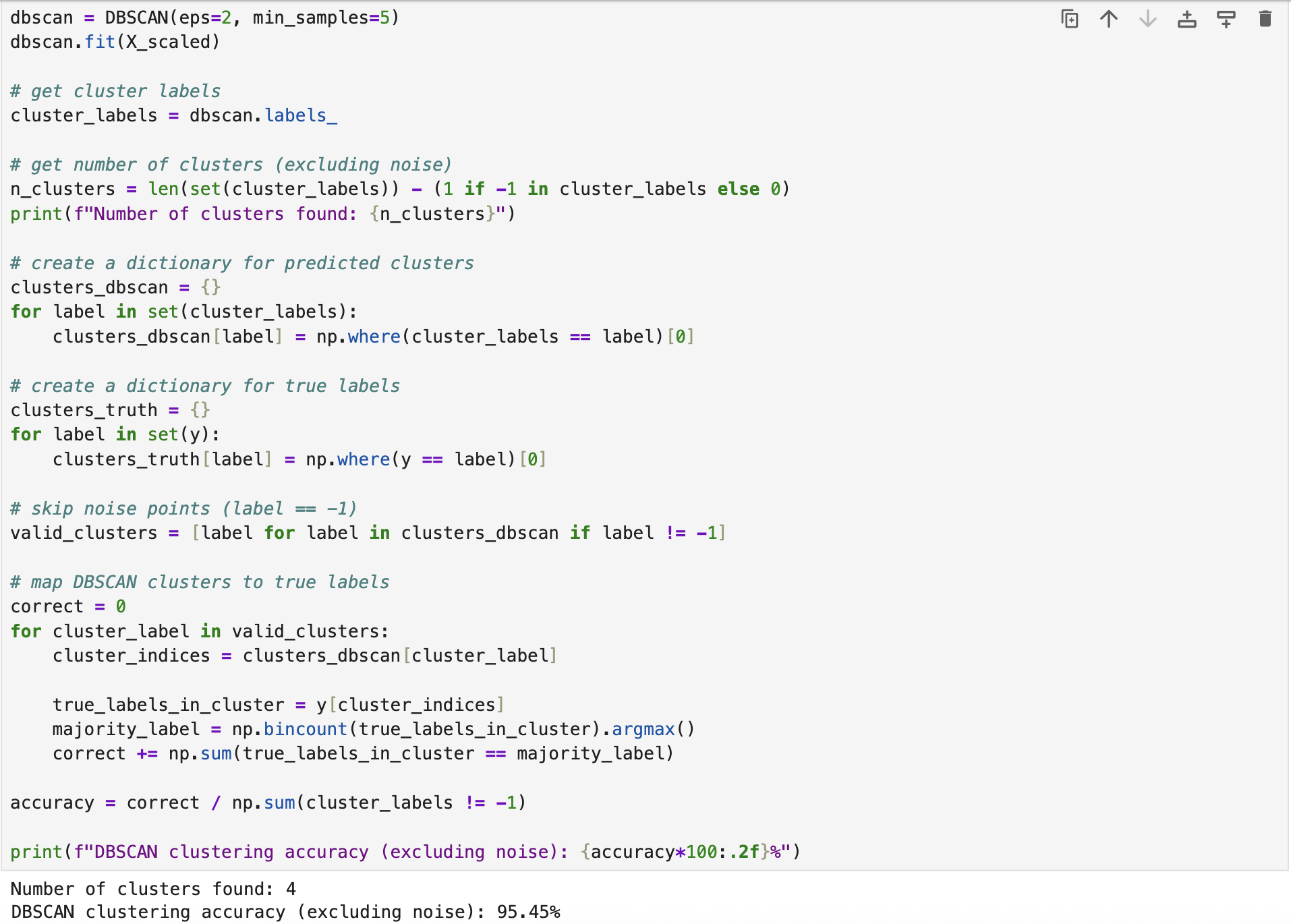
We developed a DBSCAN clustering model for the breast cancer dataset using the built-in functions provided in \textit{scikit-learn} in Python (we kept the target labels for evaluation purposes). Although DBSCAN is a powerful clustering model, it is highly sensitive to the values of eps and minPts. By default, these parameters are set to \(eps = 0.5\) and \(minPts = 5\). However, the obtained results indicate that \(eps = 4\) leads to better clustering performance.
The following screenshot illustrates the model's fitting and predicting process. The model was imported from sklearn.clusters. The complete implementation script is available on the GitHub page DBSCAN Clustering.
References
[1] F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, and E. Duchesnay, “Scikit-learn: Machine learning in Python,” Journal of Machine Learning Research, vol. 12, pp. 2825–2830, 2011.

 IEEE
IEEE Web of Science
Web of Science